I recently read “Why Scientists are Turning to Rust” and found myself disagreeing with its core premise. At same time, there is also a broader pattern in the “Python vs. Rust” claiming that Rust is the future of scientific area–for Python+C/Fortran is current optimal–discourse that fundamentally misunderstands how scientific computing actually works.
(more…)This Blog is Untitled
… and there is no description.
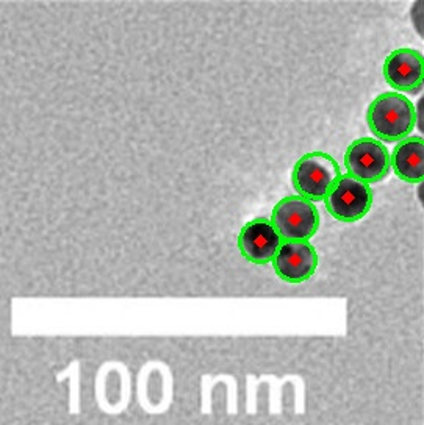
Detecting Spherical NPs from TEM
I recently read the paper “Practical Guide to Automated TEM Image Analysis for Increased Accuracy and Precision in the Measurement of Particle Size and Morphology.” In Figure 8, the spherical nanoparticles appear as faintly overlapping circles,

which immediately brought to mind the Hough Circle detection algorithm. By voting in the circle parameter space, Hough Circle can pick out weak or partially obscured arcs and thus avoid lumping overlapping particles into a single object.
Naïve Networking 2
The following note explains how to deploy a high-performance VPN using WireGuard on VPP with DPDK. This lets lab members connect to computation servers from anywhere on the campus network with minimal overhead.
(more…)Naïve Networking
The following note outlines how to set up a VPN so that lab members can connect to the computation servers from anywhere on the campus network.
1) Requirements
- Goal: Allow remote VPN clients to access lab servers on a different subnet.
- VPN software: SoftEther VPN Server (L2-bridge mode) + SoftEther VPN Client.
- Network device: Layer-3 switch that supports SVIs and inter-VLAN routing.
Unwrap big molecules & self-assemblies in PBC
Periodic boundary conditions (PBC) are ubiquitous in molecular simulations. However, when visualizing results, one often prefers to see whole molecules rather than fragments cut by the simulation box. If the molecule percolates through the box, minimizing artificial cuts becomes essential. Below are two methods for reconstructing intact structures under PBC:
(more…)