Periodic boundary conditions (PBC) are ubiquitous in molecular simulations. However, when visualizing results, one often prefers to see whole molecules rather than fragments cut by the simulation box. If the molecule percolates through the box, minimizing artificial cuts becomes essential. Below are two methods for reconstructing intact structures under PBC:
(more…)This Blog is Untitled
… and there is no description.
Vector calculus notes (1)
Problem: given a series molecules $i=1,\dots,N$, connected with junction atoms $\mathbf{x}_i^k$, for $k=1,2,…m$ if there are $m_i$ atoms from molecule $i$ that are bonded with other molecules. The connection bonds are defined as $\mathbf{b}_i := \lbrace\mathbf{x}_j^{k_1}-\mathbf{x}_i^{k_2}|\forall k_1, k_2\, \mathrm{and}\, \forall j\in \mathrm{neighbor\, of}\, i\rbrace$, assume that the center-of-mass of the molecules are fixed, but are rotatable, minimize $\sum_i |\mathbf{b}_i|^2$ with a series of rotation matrices $R_i$:
$$ R_i = \min_{R_i}(L:= \sum_i \sum_{j \in \mathrm{neighbor\, of}\, i} \sum_{k_1,k_2} |\mathrm{pbc}(R_j \mathbf{x}_{j}^{k_1} – R_i \mathbf{x}_{i}^{k_2})|^2) $$
with constraint
$$ c_1=\sum_i |R_i^TR_i – I|^2 = 0 $$
$$ c_2=\det(R_i) – 1=0 $$
the constraint ensures $R^T R =I$ and $\det(R)=1$ for each rotation matrix thus $R_i$ are rotation matrices. The periodic boundary condition is introduced for most simulation systems.
Minimization procedure includes calculation of jacobian of loss and constraint functions:
$$ \frac{\partial L}{\partial R_i} = -2 \sum_{j \in \mathrm{neighbor\, of}\, i} \sum_{k_1, k_2} \mathrm{pbc}(R_{j} \mathbf{x}_{j}^{k_1} – R_i \mathbf{x}_i^{k_2})^T \mathbf{x}_i^{k_2} $$
and
$$\frac{\partial c_1}{\partial R_i} = 4 (R_i R_i^T – I) R_i $$
$$\frac{\partial c_2}{\partial R_i} = \det(R_i) (R_i^{-1})^T $$
Or the merged constraint (?):
$$ c = \sum_i |R_i^TR_i – I|^2 + (\det (R_i)-1)^2 = 0 $$
with
$$\frac{\partial c}{\partial R_i} = 4 (R_i R_i^T – I) R_i + 2(\det(R_i)-1)\det(R_i)(R_i^{-1})^T $$
def cons_jac(R): # Only the second part
R = R.reshape(-1, 3, 3)
det = np.linalg.det(R)
inv = np.linalg.pinv(R)
return 2 * det[:, None, None] * (det[:, None, None] - 1) * np.swapaxes(inv, (1, 2))
Calculating SQ with FFT
The structure factor, commonly denoted as $S(\mathbf{q})$, is an essential characterization quantity in molecular simulations. Its calculation is straightforward based on its definition. Since periodic boundary conditions are typically used in these simulations, we have
$$S(\mathbf{q}) := \mathcal{FT}\lbrace\langle\rho(\mathbf{r})\rho(\mathbf{0})\rangle\rbrace(\mathbf{q})$$
which actually represents a circular convolution. This means that using the Fast Fourier Transform (FFT) can dramatically accelerate the computation compared to performing the calculation directly.
The FFT-based procedure proceeds as follows:
- Compute the density field $\rho(\mathbf{r})$, which is a sum of Dirac delta functions approximated by a histogram.
- Use the FFT to calculate the Fourier transform $\hat{\rho}(\mathbf{q})$.
- Obtain the structure factor by taking the modulus squared of the transform:
$$ S(\mathbf{q}) = |\hat{\rho}(\mathbf{q})|^2. $$ - Average over the modulus to yield
$$ S(q) = \frac{\int S(\mathbf{q})\delta(|\mathbf{q}|-q)\,\mathrm{d}\mathbf{q}}{\int\delta(|\mathbf{q}|-q)\,\mathrm{d}\mathbf{q}}. $$
In terms of efficiency, if the simulation box is divided into $N$ bins (for example, in 3D, $N = N_x N_y N_z$), the FFT has a computational complexity of $O(N\log(N))$, and the averaging step is $O(N)$. By contrast, a direct method would involve a loop over all particles and $N$ bins in $\mathbf{q}$ space—and typically, the number of particles is much larger than $\log(N)$. Moreover, because most simulation data are real-valued, employing an FFT designed for real inputs (rFFT) can further improve performance.
Below is an example code for a 3D case, where the indices i, j, and k correspond to $N_x$, $N_y$, and $N_z$, respectively.
fftn(a) == np.concatenate([rfftn(a), conj(rfftn(a))[-np.arange(i),-np.arange(j),np.arange(k-k//2-1,0,-1)]], axis=-1)Accuracy Considerations: Effect of Bin Size
The histogram bin size should be smaller than half the minimum pairwise distance in the system, in accordance with the Nyquist-Shannon sampling theorem. For instance, in Lennard-Jones systems, a bin size of approximately $0.4\sigma$ is advisable. However, if you are only interested in small $q$ values—as is often the case in self-assembly systems—full pairwise resolution is not necessary; a sampling interval that is just smaller than half of the domain of interest may suffice.
Randomness Within Bins
Particles are assumed to be randomly distributed within each bin, especially when employing large bins. The Fourier transform of the particle positions is given by
$$\sum_i \exp(-\mathrm{i}\mathbf{q}\cdot\mathbf{r}_i)$$
This sum can be decomposed as
$$\sum_\mathrm{bin} \exp(-\mathrm{i}\mathbf{q}\cdot\mathbf{r}_\mathrm{bin})\left(\sum_i \exp(-\mathrm{i}\mathbf{q}\cdot\delta\mathbf{r}_i)\right)$$
where each particle’s position is written as
$$\mathbf{r}_i = \mathbf{r}_\mathrm{bin} + \delta\mathbf{r}_i$$
Assuming the displacements $\delta\mathbf{r}_i$ are randomly distributed within the bin, the inner sum can be approximated by
$$n_\mathrm{bin}\langle\exp(-\mathrm{i}\mathbf{q}\cdot\delta\mathbf{r})\rangle$$
with the average taken over the distribution of $\delta\mathbf{r}$. For a one-dimensional example, if $\delta r$ is uniformly distributed in the interval $(-L/2, L/2)$ (where $L$ is the bin size), the average is given by $\mathrm{sinc}(qL/2)$. Multiplying by this sinc correction during step 4 yields a properly corrected $S(q)$.
This strategy ensures that both efficiency and accuracy are maintained when computing the structure factor in molecular simulations.
Draw paths on energy landscape
A path on energy landscape is like:
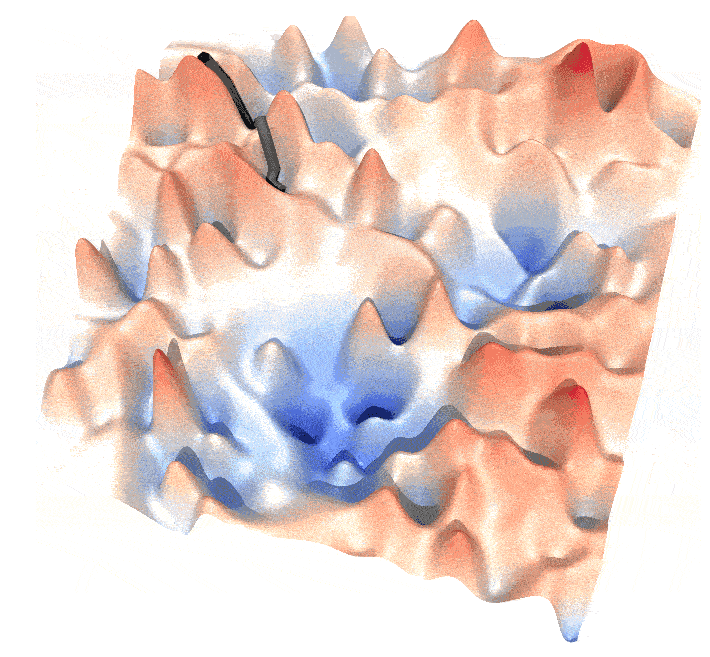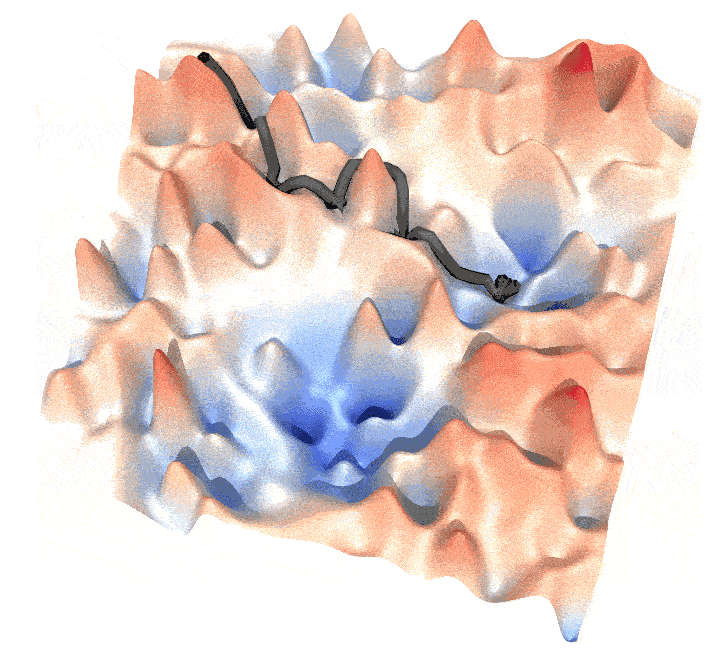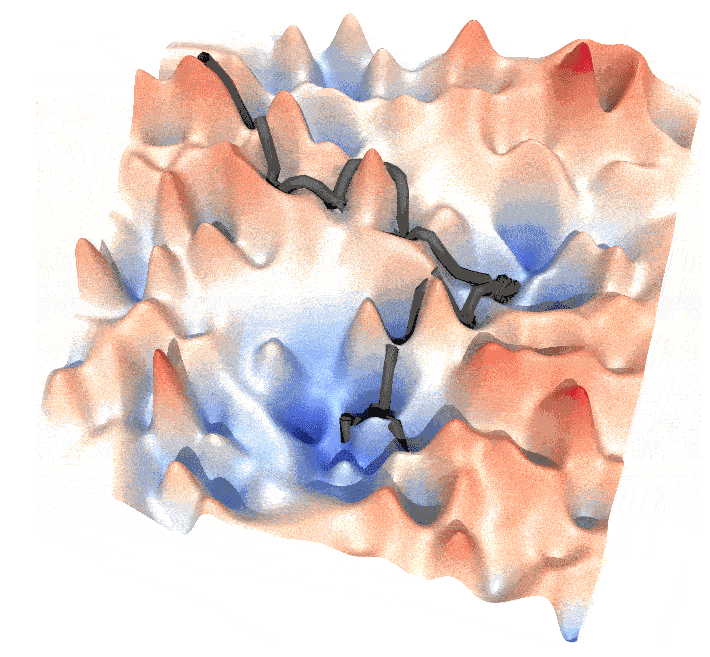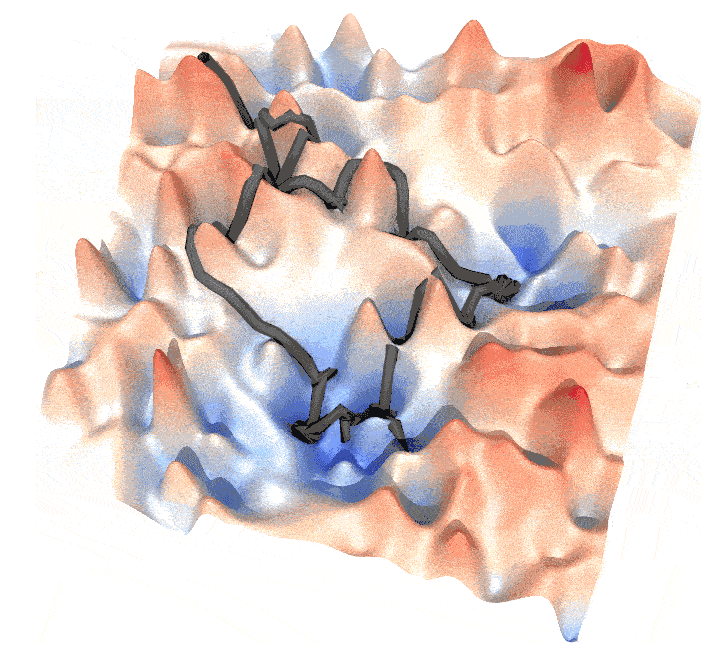
and this post is a guide to generate such animations.
Step 1: Generate the Energy Landscape
An energy landscape is essentially an energy function $U(\mathbf{x})$. This function can be created in various ways. For example, you might generate a series of Gaussian functions with random parameters $\mu_i$, $\sigma_i$, and weight factors $\omega_i$. From these, you can construct a probability function $P(\mathbf{x}) = \sum_{i=1}^N \omega_i G(\mathbf{x}; \mu_i, \sigma_i)$. The energy function $U(\mathbf{x})$ is then derived using Boltzmann inversion: $U = -k_B T \log P$ .
In the example above, I generated a series of Mexican hat functions with varying parameters ($a$), ($b$), and locations.
Step 2: Generate the Path
A path is determined by the dynamics of the system, such as $\dot{\mathbf{x}} = -\nabla U(\mathbf{x})$ or $\ddot{\mathbf{x}} – \dot{\mathbf{x}} = -\nabla U(\mathbf{x})$. By integrating from a starting point, you can generate a path. In our example, the walker follows a cyclic path, which requires an understanding of the Lagrangian.
A walker on an energy surface from point $A$ to $B$ takes the “canonical path” where the Lagrangian is minimized. The probability of the walker being at $A$ at time $t_0$ and reaching $B$ at time $t_1$ is given by:
$$
P(\mathbf{x}_A, t_0 \mid \mathbf{x}_B, t_1) = \frac{1}{Z} \int_{\mathbf{x}(t_0) = \mathbf{x}_A}^{\mathbf{x}(t_1) = \mathbf{x}_B} \mathcal{D}\mathbf{x} \exp\left(-\int_{t_0}^{t_1} L(\mathbf{x}, \dot{\mathbf{x}}, t) \, dt\right)
$$
Maximizing this probability implies minimizing the Lagrangian $L$ (saddle point approximation). Given $L = T – U$ with $T \propto \dot{\mathbf{x}}^2$, we can discretize the integral of $L$ as:
$$
L = \sum_{i=0}^N \lambda_1 (\mathbf{x}_{i+1} – \mathbf{x}_i)^2 – U(\mathbf{x}_i)
$$
where $\mathbf{x}_0 = \mathbf{x}_A$ and $\mathbf{x}_N = \mathbf{x}_B$. Here, $\lambda_1$ is a hyperparameter controlling the path’s tightness. Our goal is to find a series of $\mathbf{x}_i$ that minimizes $L$. This can be achieved using scipy.optimize.minimize
Step 3. Generate the animiation
The output can be drawn by mayavi, in the example I draw the frames of the animation by povray, where .pov files were generated by mayavi. The trajectory is a thick line that slightly shifted higher than the energy landscape surface to ensure a clear view. You can also draw a big point (sphere) at the forefront of a trajectory, to emphasize the walker.
The mighty RDKit
Mapping coarse-grained models back to all-atom models is a tricky problem. E.g., for polymeric systems, CG models are often used to polymerize monomers to cross-linked polymer networks. Mapping back these systems to all-atom models need to handle reactions amongst monomers. Dealing back-mapping case by case is dumb: for one specific kind of reactions, there are a family of polymers, i.e., the reaction of Polyethylene could also be used for Polystyrene.
Brief introduction to SMARTS
SMARTS is a language for describing molecular patterns, one may find details from this link. SMARTS is not only aimed to search substructures from a molecule, it could also define abstractly a reaction in the way of functional groups, for example:
'[C:1](=[O:2])O.[N:3]>>[C:1](=[O:2])[N:3]'defines an amidation reaction, any RCOOH and RNH2 are connected as RCO-NHR. Therefore, we may use SMARTS to match the functional groups in reactants and build productions from the pre-defined rules.
RDKit: Open-Source Cheminformatics Software
Luckily, we needn’t to handle SMARTS by hand, the RDKit offers convenient tools dealing with reactions. There is an example from RDKit:
from rdkit import Chem
from rdkit.Chem import rdChemReactions
rxn = rdChemReactions.ReactionFromSmarts('[C:1](=[O:2])O.[N:3]>>[C:1](=[O:2])[N:3]')
reacts = (Chem.MolFromSmiles('C(=O)O'),Chem.MolFromSmiles('CNC'))
products = rxn.RunReactants(reacts)
len(products)
1
len(products[0])
1
Chem.MolToSmiles(products[0][0])
'CN(C)C=O'we can see that RDKit compiles the SMARTS reaction rule and applied it on reactants by rxn.RunReactants method. Notably, the number of outcomes is not necessarily 1, for C-C + C-C -> C-C-C-C reaction, there are 4 possible outcomes, which are permutations of C11-C12-C21-C22, C11-C12-C22-C21, …, we will use this property in the following work.
Situation: Making huge molecules
For huge molecules (polymers for example), we of course may initiate reactions naturally, e.g., for chain growth, we can code as p=rxn.RunReactants([p, monomer]). However, for large molecules (with 10000+ atoms), this is very inefficient. Especially for cross-linked polymer network, since cross link may happen on any available sites, the number of products given by RDKit may be explosively large. Moreover, in case of back-mapping, the topology of CG model is fixed, i.e., reactions on CG level are already done. It is very dumb that we filtrate out from all possible products instead of directly “react” on the given sites, so we need to react “locally” according to the CG topology. Now the problem is that we cannot actually let the 2 active monomers “react”, for the polymers grow stepwisely: for instance, a polymer ABC may grow from following sequences, A+B->AB, AB+C->ABC, or B+C->BC, BC+A->ABC. The “element” reactions (or connection rules of monomers to be more precise) are A+B and B+C, if A and B are choosen first, after reaction, reactants become products, AB. In the next reaction, B and C, we’ll have to handle AB+C rather than B+C. To avoid troubles like this, we have to keep the monomer un-reacted and only trace the changes, then apply all changes at one-time after all reactions. We therefore need to make a map which maps atoms in products to reactants.
Atom and bond maps
Atom map
In any reaction, atoms are classified into 2 categories, reacting atoms and un-reacting atoms. Here I give an example to map atoms between products and reactants after rxn.RunReactants. We first need to preprocess the reactants, in RDKit, we can use atom.SetIntProp to assign an integer property to atom, in this case, if we have multiple reactants, i.e., (r1, r2, r3,…)->(p1, p2, p3,…), we will add property “reactant_idx=1,2,3,..” to atoms in (r1, r2, r3,…) accordingly.
We also need a container to record information of an atom: for an atom in one product (product_id and product_atom_id), we need to known where the atom come from (reactant_id), which atom of the corresponding reactant (reactant_atom_id), for example, for reaction ‘A-B+C-D->A-B-D+C’, atom D has product_id=0, reactant_id=1, reactant_atom_id=1, product_atom_id=2. The codes are shown blow, property “reactant_idx” set for every atom in RDKit.Chem.Mol helps to map un-reacted atoms.
Bond map
The bond map is also straightforward, we need to know a bond (i, j, bond_type) from production and where its atoms came from. In a reaction, a bond in reactant may change its bond type, e.g., double bond to single bond, or break. Also bonds in production may generate, as in former example, bond B-D.
atom_info = namedtuple('atom_info',
('reactant_id', 'reactant_atom_id', 'product_id', 'product_atom_id', 'atomic_number'))
bond_info = namedtuple('bond_info',
('product_id', 'product_atoms_id', 'reactants_id', 'reactant_atoms_id', 'bond_type', 'status'))
def process_reactants(reactants):
r"""Give reactant index for reactants, please do not use SAME molecules, e.g., [m, m]
:param reactants: list of reactants
:return: list of reactants
"""
for r_idx, m in enumerate(reactants):
for atom in m.GetAtoms():
atom.SetIntProp('reactant_idx', r_idx)
return reactants
def map_reacting_atoms(reaction):
r"""
:param reaction: Reaction object
:return: dict, reacting atoms: reactant index
"""
reacting_map = {}
for r_idx in range(reaction.GetNumReactantTemplates()):
# print(r_idx, reaction.GetNumReactantTemplates())
rt = reaction.GetReactantTemplate(r_idx)
for atom in rt.GetAtoms():
if atom.GetAtomMapNum():
reacting_map[atom.GetAtomMapNum()] = r_idx
return reacting_map
def map_atoms(products, reacting_map):
r"""
:param products: list of products
:param reacting_map: reacting atom map
:return: atom map and reacting atoms
"""
amap = []
ra_dict = {}
for ip, p in enumerate(products):
p_idx = ip
for a in p.GetAtoms():
p_aidx = a.GetIdx()
old_mapno = a.GetPropsAsDict().get('old_mapno')
r_aidx = a.GetPropsAsDict().get("react_atom_idx")
if old_mapno is not None: # reacting atoms
r_idx = reacting_map[old_mapno]
if ra_dict.get(r_idx) is None:
ra_dict[r_idx] = []
ra_dict[r_idx].append(r_aidx)
else:
r_idx = a.GetPropsAsDict().get('reactant_idx')
amap.append(atom_info(r_idx, r_aidx, p_idx, p_aidx, a.GetAtomicNum())) # all Atoms
return amap, ra_dict
def atom_map(products, reaction):
r"""Map atoms between reactants and products
:param products: product list
:param reaction: reaction object
:return: atom map and reacting atoms
"""
reacting_map = map_reacting_atoms(reaction)
amap, reacting_atoms = map_atoms(products, reacting_map)
return amap, reacting_atoms
def bond_map(reactants, products, reaction):
r"""Map bonds between reactants and products
:param reactants: list of reactants
:param products: list of products
:param reaction: reaction object
:return: list of bond map
"""
amap, reacting_atoms = atom_map(products, reaction)
res = []
for ir, r in enumerate(reactants):
for bond in r.GetBonds(): # exist in reactants, but not in production
a, b, t = bond.GetBeginAtomIdx(), bond.GetEndAtomIdx(), bond.GetBondType()
for atom in amap:
if atom.reactant_id == ir:
if atom.reactant_atom_id == a:
a_pid = atom.product_id
a_paid = atom.product_atom_id
if atom.reactant_atom_id == b:
b_pid = atom.product_id
b_paid = atom.product_atom_id
if a_pid == b_pid:
p_bond = products[a_pid].GetBondBetweenAtoms(a_paid, b_paid)
if p_bond is None:
res.append(bond_info(a_pid, (a_paid, b_paid), (ir, ir), (a, b), t, 'deleted'))
for ip, p in enumerate(products):
for bond in p.GetBonds(): # exist in productions, compare with reactants
a, b, t = bond.GetBeginAtomIdx(), bond.GetEndAtomIdx(), bond.GetBondType()
for atom in amap:
if atom.product_id == ip:
if atom.product_atom_id == a:
a_rid = atom.reactant_id
a_raid = atom.reactant_atom_id
if atom.product_atom_id == b:
b_rid = atom.reactant_id
b_raid = atom.reactant_atom_id
if a_rid != b_rid:
res.append(bond_info(ip, (a, b), (a_rid, b_rid), (a_raid, b_raid), t, 'new'))
if a_rid == b_rid:
r_bond = reactants[a_rid].GetBondBetweenAtoms(a_raid, b_raid)
if r_bond is None:
res.append(bond_info(ip, (a, b), (a_rid, b_rid), (a_raid, b_raid), t, 'new'))
else:
if r_bond.GetBondType() != t:
res.append(bond_info(ip, (a, b), (a_rid, b_rid), (a_raid, b_raid), t, 'changed'))
return resExmaple: Making polymers
2-monomer reactions
If the system only contains 2-monomer reactions, for example, for PS or PE, we only have A-A where the reactions always take form as P+M->PM. For some cross-linking system, a branch site B on the chain, ..A-B-A…, if B has a cross-linking functional group which reacts with C, i.e., …A-B(C)-A…, the reaction is …A-B-A… + C -> …A-B(C)-A…., the reaction are actually happened on B+C. For such cases, we only need to iterate over the edges (CG polymer is a GRAPH made from conjunctions (EDGES) of monomers (NODES)) and using RDKit to handle the reactions between the 2 active monomers.
Complex reactions involve 3 or more monomers and multi-step reactions
For example, melamine. Where 3 molecules react into an aromatic ring. And phenol (A) formaldehyde (B) resins, the formaldehyde (B) changes after first step reaction with phenol (A), the actual reaction is a 2-step reaction: A+B->AB, AB+A->ABA. For these 2 cases, we need more detailed information in CG models, the reactions and corresponding SMARTS should be defined as an overall reaction, e.g., A+B+A->ABA.
- Get monomers by monomer index;
- Apply reaction rules and get atom and bond maps;
- Record the changing of atoms and bonds;
- Record which atoms are involved in this reaction;
Step.4 is important, remember that we don’t allow actual reactions happen, for every reaction, we all take the original molecules to run the reactions (if there are 2-or-more-step reactions, simply add them up). For some symmetrical monomers, e.g., H2N-Ar-NH2, which has 2 same sites. If one site has already reacted, then this site is forbidden in the next reaction. There are at least 4 possible outcomes for 2 symmetrical monomers, we need to iterate over these 4 outcomes to pick out the reaction with un-reacted functional groups for each monomer.
Force field
Force field parameters of a given atom is defined from its chemical environment. After back-mapping of the topology, we can further use Foyer or openforcefield to generate force field parameters, both Foyer and openforcefield can easily get topology from RDKit. Or we can simply use rdkit.Chem.rdMolChemicalFeatures model to find parameters from pre-defined force field database, e.g., “PARAMETER” “CHEMICAL ENV SMARTS” or compare molecule similarities from database.
